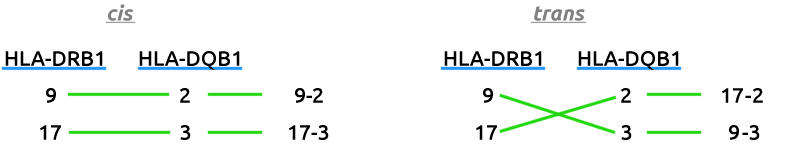|
Where can I obtain testing?
|
| |
|
Tests can be ordered by any practitioner and in many cases are covered by insurance. Direct to consumer options exist in the United States that do not require a prescription. If no options exist in the table below for your region or no test code is listed, request testing at any capable laboratory and ensure at minimum typing is performed for the genes HLA-DRB1 and HLA-DQB1. Typing for the genes HLA-DRB3, HLA-DRB4 and HLA-DRB5 should always be performed as well if possible.
|
| |
|
|
| |
|
Direct to consumer testing offered by Life Extension and True Health Labs is identical to testing offered by Labcorp and draws are performed at Labcorp locations. All costs listed are costs without insurance coverage, though with the exception of the direct to consumer options which do not require a doctor, prescription or insurance, tests are usually covered.
|
| |
|
If you know of additional testing options either within these regions or in regions not listed, speak up so they can be added to this table.
|
|
I thought that I had two haplotypes, one from mom and one from dad, however the calculator has shown me more than two. How many haplotypes do I have?
|
| |
|
When referring to haplotypes whose alleles were inherited from the same parent, there are two and only two. One comes from an individual's mother and the other from their father. These are the two haplotypes the HLA-DR Calculator will display in the vast majority of cases.
|
| |
|
That being said, when referring to HLA-DR/DQ haplotype combinations as we are, you can technically have up to four using two different methods of isoform pairing1. Two are considered in cis, and two in trans. Your two cishaplotypes will combine mom's alleles with mom's and dad's with dad's, whereas your transhaplotypes will pair one of mom's with one of dad's.
|
| |
|
| |
|
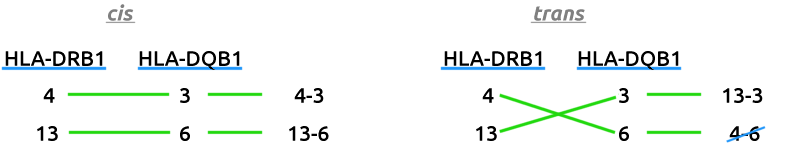
The third figure has been omitted for simplicity, as it is always associated with and can be derived from the first.
|
| |
|
In the absence of having phased data, where alleles from the same parent are clearly identified and labeled, Dr. Shoemaker's rosetta stone process attempts to phase the data via process of elimination. In the example above, one method of combining yields two haplotypes that exist, whereas the other yields only one. In this very clear case, you would have 13-6 and 4-3 in cis and 13-3 in trans, as 4-6 does not exist.
|
| |
|
Some cases are ambiguous and phase can not be determined with this method, and it is therefore not possible to determine which set's constituents were inherited from the same parent, and therefore which haplotypes are in cis and which are in trans.
|
| |
|
Take the example below:
|
| |
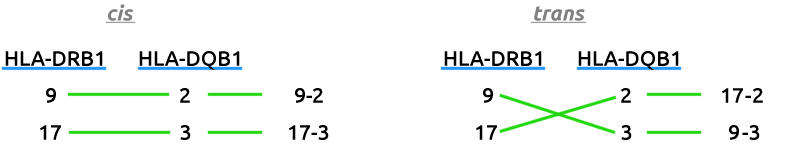
|
| |
|
Both sets exist when combining both ways, and it is therefore not possible to determine which two haplotypes are in cis and which are in trans.
|
| |
|
As of version 1.2 and later, the HLA-DR calculator will show your two cishaplotypes only within the main results. Transhaplotypes, if any are detected, will be discussed separately on the details page. The exception is an ambiguous condition, in which case both possible sets are shown. There is no scientific consensus on the relevancy of transhaplotypes, however several papers do reference them and Dr. Shoemaker's rosetta stone process does consider susceptible haplotypes relevant even if present in trans. For this reason, they are mentioned.
|
| |
|
I've already mapped my genome via 23andMe or Ancestry.com, can I somehow decode my HLA-DR haplotype from the raw data?
|
| |
|
No
|
| |
|
But 23andMe reads my genes, and my HLA-DR haplotype is genetic. They both involve genes. The answer MUST be in there!
|
| |
|
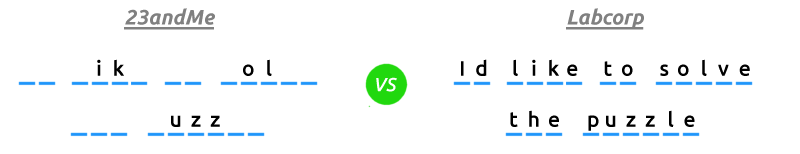
A common misconception is that saliva based genotyping services such as 23andMe map your entire genome. They do not unfortunately. 23andMe genotypes around 600,000 loci out of the approximately 50,000,000 that comprise the human genome. This is simply not enough data to compute a haplotype for HLA-DRB1 or HLA-DQB1. If you would like to have your full exome sequenced, with all 50 million loci included in your raw data, that can be performed by a company called Genos Research for approximately $499. With that data it should be possible, otherwise sequenced based typing via a blood test available at Labcorp, Quest and other laboratories will be necessary.
|
| |
|
| |
|
But I ran my raw data through livewello or mthfrsupport and I can see mutations for HLA genes. This is a puzzle BEGGING to be solved. The answer must be in there!
|
| |
|
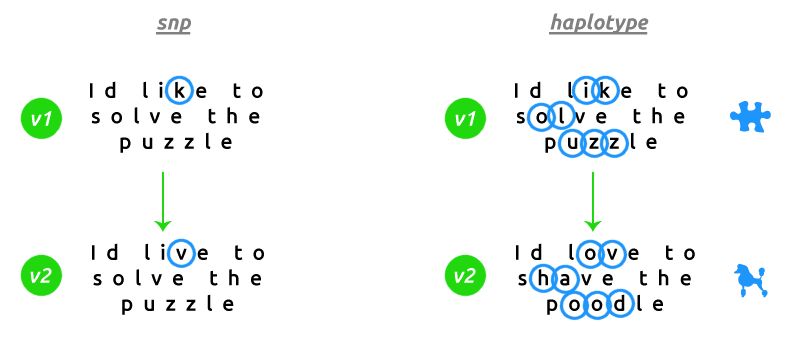
First it's important to make the distinction between a single nucleotide polymorphism (SNP) and a haplotype. A SNP is a change in one letter of a gene. MTHFR C677T exchanges a C for a T at position 677 on the gene MTHFR. That's all. A haplotype however is a pattern consisting of multiple SNPs on a single gene. There may be 40 different SNPs that comprise the haplotype HLA-DRB1*01. That's haplotype version 1 on the gene HLA-DRB1. If you think of a gene as a sentence, a SNP changes only one letter of that sentence wheras a haplotype will change several letters creating an entirely different version of that sentence.
|
| |
|
| |
|
Services such as Livewello and MthfrSupport may report handful of SNPs on the gene HLA-DRB1, but far from enough to compile a haplotype for that gene alone. In addition to that, your complete HLA-DR/DQ haplotype is comprised of individual haplotypes (each containing multiple SNPs) on up to five genes: HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5 and HLA-DQB1; and 23andMe raw data contains ZERO information for the genes HLA-DRB3 and HLA-DRB4.
|
| |
|
It's the difference between individual letters, words and sentences. 23andMe is working with individual letters, while your HLA-DR haplotype is written in full sentences.
|
| |
|
I see references made to versions of my haplotype. What does this mean?
|
| |
|
When we are tested for biotoxin susceptibility we have our haplotypes read at low resolution, or at a length of two digits. When it comes to disease risk and disease protection some research may refer to high resolution haplotypes, or those at a length of four digits.
|
| |
As an example, if your value for HLA-DRB1 is 04 when read at low resolution, you may have a value of 0401, 0402, 0403, 0404, 0405 or many others. Disease risk and disease protection may be correlated with some versions, but not others. To determine whether you possess the specific haplotype version referenced in the cited literature high resolution testing is recommended, which is offered by both Quest and Labcorp.
© Labcorp High Resolution HLA-DRB1 - Test Code 167167
© Labcorp High Resolution HLA-DQB1 - Test Code 167232
© Quest High Resolution HLA-DRB1 - Test Code 17393
© Quest High Resolution HLA-DQB1 - Test Code 17394
|
| |
|
High resolution testing is available for HLA-DRB3, HLA-DRB4 and HLA-DRB5 however I have yet to see relevant literature refer to high resoltion haplotypes on those genes.
|
| |
|
I notice there is an Australian HLA-DR calculator, how does this differ from the USA Labcorp version?
|
| |
|
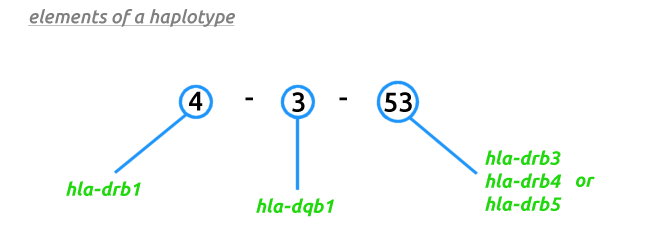
Your HLA-DR/DQ haplotype is comprised of sometimes two, but most often three figures (see the question on elements of a haplotype for a detailed explanation). In the United States Labcorp, Quest and possibly other labs perform sequence based typing on all genes involved (HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5 and HLA-DQB1) providing data for all three figures. In Australia, Sonic Healthcare and NutriPATH return haplotypes for only the first two figures (HLA-DRB1 and HLA-DQB1). The Australian HLA-DR calculator has been designed to show all possibilities given the missing third figure.
|
| |
|
Additionally, as of version 1.2 the Australian HLA-DR calculator is able to leverage serology and prevalence to derive or significantly narrow down the third figure to the extent that biotoxin susceptibility can be ascertained with a high degree of certainty in 94% of cases.
|
| |
|
How does the Australian HLA-DR Calculator use serology to derive the missing third figure?
|
| |
|
Certain genetic variations are linked as part of what is known as a serotype, and the existence of some genes is dependent on the version of another. The first and third figure of your HLA-DR/DQ haplotype behave in this fashion.
|
| |
|
When the value of HLA-DRB1 is a 4, 7 or 9 the presence of HLA-DRB4 can be inferred resulting in a third figure value of 53.
|
| |

|
| |
|
When the value of HLA-DRB1 is a 15 or 16, the presence of HLA-DRB5 can be inferred resulting in a third figure value of 51.
|
| |

|
| |
|
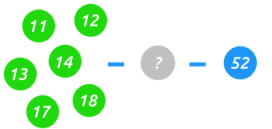
When the value of HLA-DRB1 is an 11, 12, 13, 14, 17 or 18, the presence of HLA-DRB3 can be inferred resulting in a partial third figure value of 52.
|
| |
|
| |
|
In the case of HLA-DRB3 there are three common haplotypes, 01, 02 and 03 leading to a third figure value of 52A, 52B and 52C respectively. Using serology the presence of HLA-DRB3 can be inferred, but not the specific haplotype. While we can't know the exact haplotype definitively, in some cases we can leverage prevalence to narrow down our possibilities, often with high probability.
|
| |
|
In cases where HLA-DRB1 is an 11, 12 or 14 the most frequent HLA-DRB3 haplotype associated is 02, leading to a third figure of 52B. Importantly, HLA-DRB3 haplotypes 01 and 03 (leading to a third figure of 52A and 52C respectively) do in fact exist and occur in conjunction with HLA-DRB1 haplotypes 11, 12 and 14, however they are exceedingly rare. For this reason, a third figure of 52B can be predicted with high probability.
|
| |
|
In cases where HLA-DRB1 is a 13, all possibilities lead to the same biotoxin susceptiblilty and the individual haplotype for HLA-DRB3 is inconsequential. 13-3-52A and 13-3-52B are both multisusceptible, 13-6-52A, 13-6-52B and 13-6-52C are all mould susceptible and 13-7-52A, 13-7-52B and 13-7-52C are all benign.
|
| |
|
When HLA-DRB1 is a 17, this is a special case, and the only case where biotoxin susceptibility can not be inferred with high probability. This is a case where the individual haplotype for HLA-DRB3 matters. 17-2-52A and 17-2-52B are both mould susceptible, while 17-2-52C is benign. This leaves a 66% probability of a mould susceptible haplotype and a 33% probability of a benign with no way to narrow down any further. All other haplotypes involving an HLA-DRB1 value of 17 are benign.
|
| |
|
It's important to note that while these methods can infer, derive or significantly narrow down values for the missing third figure, they are no substitute for having HLA-DRB3, HLA-DRB4 and HLA-DRB5 sequenced for certainty. Genetic anomalies and abnormalities do occur, and statistically some will fall into these categories.
|
| |